Version 4.4
Dengue Virus Typing Tool
Introduction
Introduction
This tool provides four levels of classification of dengue virus: serotype, genotype, major lineage and minor lineage. The serotype is determined based on a blast analysis against the reference sequences for the different serotypes. The genotype, major lineage and minor lineage are determined based on phylogenetic analysis.
The definition of a dengue genotype is based on a diversity of variables, e.g., monophyly, pairwise distance within and between groups, net genetic diversity within groups, etc. For each genotype reference sequences were carefully selected to represent its diversity. In addition, extensive testing was performed to be sure that our reference strains accurately classify other sequences. Since tool version 4.0 the genotypes have been slightly expanded by pulling back the defining node closer to the root, so that less sequences are unassigned.
- The serotypes are: 1, 2, 3 and 4.
- Serotype 1 includes 1I ,1II, 1III, 1IV, 1V and 1VI genotypes, and a newly defined 1VII genotype.
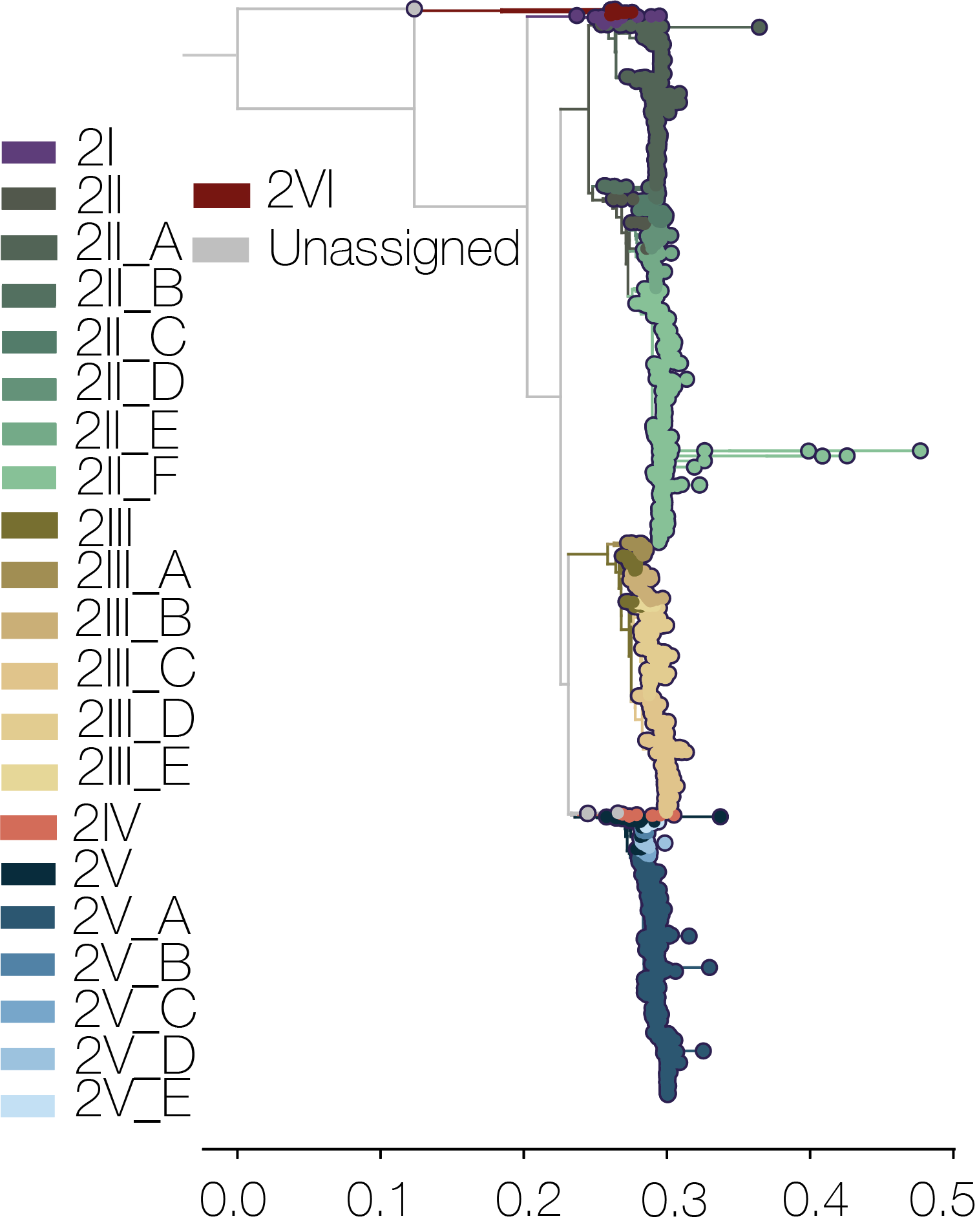
- Serotype 2 includes 2I, 2II, 2III, 2IV, 2V and 2VI genotypes.
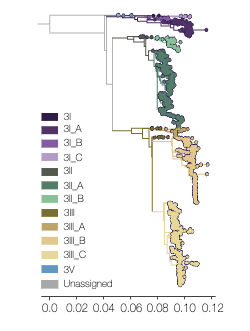
- Serotype 3 includes 3I, 3II, 3III and 3V genotypes.
- Serotype 4 includes 4I, 4II, 4III and 4IV genotypes.
In addition to the genotypes, two additional layers of classification (major and minor lineage) have been added to the tool, as proposed in Hill et al. (2024). Major and minor lineages are based on phylogenetic distance, monophyly, and the size of the clade. There are a handful of major lineages in most (not all) genotypes, denoted using latin letters, e.g. 2II_A is major lineage A in genotype II serotype 2. I and V are skipped to avoid confusion because these are used in genotype designations. There are also minor lineages for some of the major lineages, denoted by dots and numbers (e.g. 1II_A.1.2). Importantly these lineages are neutral descriptors of diversity, and aim to provide additional temporal and spatial granularity to the spread of dengue virus globally, as well as standardise the discussion of important diversity.
- Serotype 1
- Genotype I: A, B, C, D, E(1.1, 1.2, 2, 3, 4), F, G, H(1-3), J, K(1.1, 1.2, 2, 3, 4, 5, 6, 7)
- Genotype III: A(1-4), B
- Genotype IV: A, B(1-2), C
- Genotype V: A, B, C, D(1.1, 1.2, 2), E(1-4), F, G
- Genotype VII: A, B
- Serotype 2
- Genotype II: A(1.1.1, 1.1.2, 1.2, 2.1, 2.2), B, C(1-2), D(1.1, 1.2, 2, 3), E(1-2), F(1.1.1, 1.1.2, 1.1.3, 1.1.4, 1.1.5, 1.1.6, 1.2, 1.3, 2.1, 2.2)
- Genotype III: A(1-2), B, C(1.1, 1.2, 2), D(1.1, 1.2, 1.3, 2, 3), E
- Genotype V: A(1.1, 1.2, 1.3, 2, 3, 4), B, C, D, E
- Serotype 3
- Genotype I: A(1.1, 1.2, 2), B, C
- Genotype II: A(1-5), B
- Genotype III: A(1-2), B(1, 2, 3.1, 3.2), C(1, 2.1, 2.2)
- Serotype 4
- Genotype I: A(1.1, 1.2, 2, 3), B(1-2)
- Genotype II: A(1-2), B(1.1, 1.2, 2)
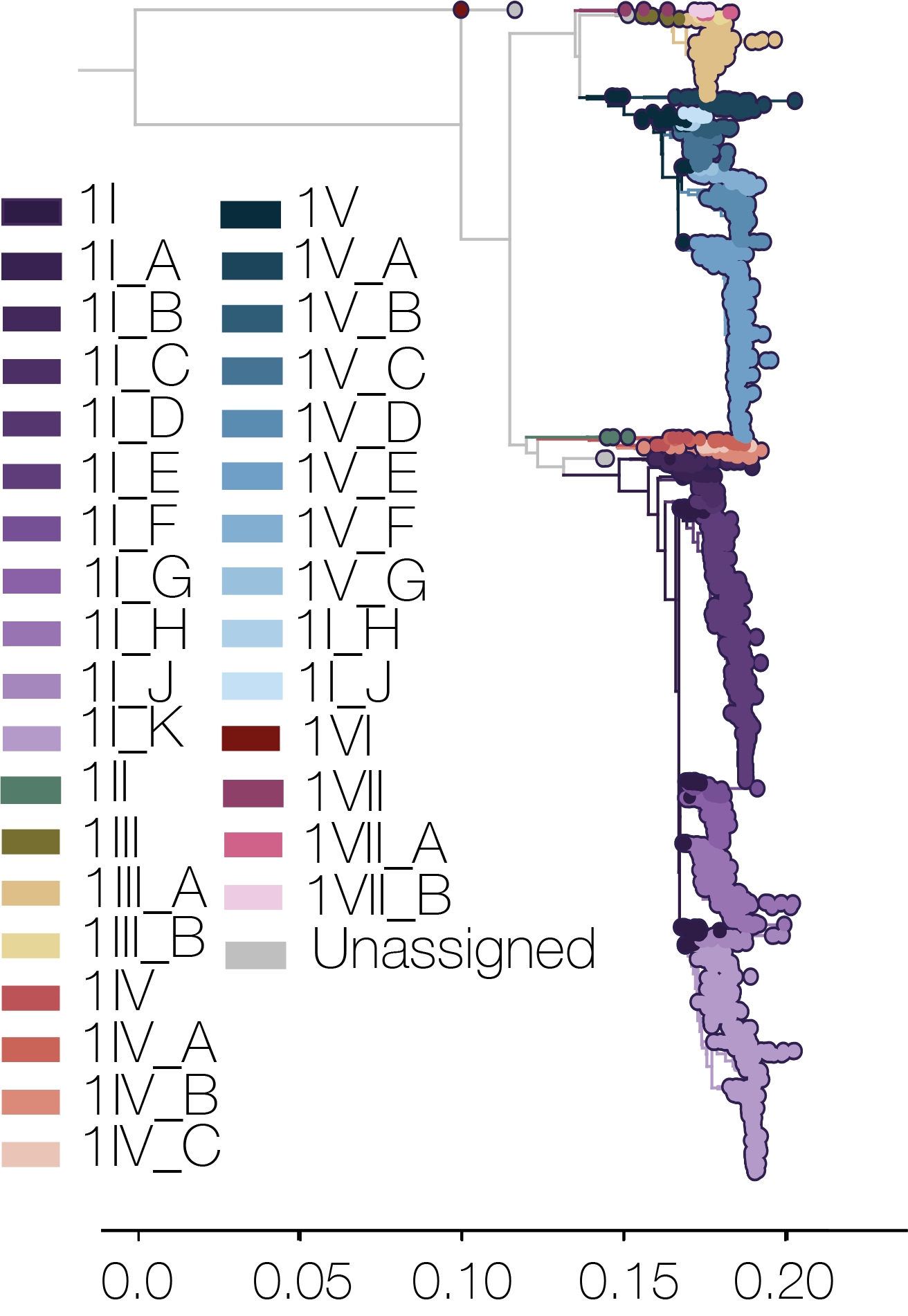
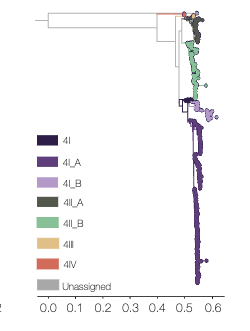
This lineage system has been a wide global collaborative effort, and so we thank everyone who has provided input, feedback and data.
There is broad consensus that the envelope glycoprotein gene (E) is suitable for phylogenetic classification, since it contains sufficient phylogenetic signal (1,485bp) to distinguish dengue from other viruses and differentiate between serotypes and genotypes. Phylogenetic trees inferred with E gene sequence data generate topologies very similar to whole genome trees. Therefore, investigators working with dengue sequences can expect similar classification accuracy when using either WGS or E gene for both genotype and major lineage.
In past versions of the Genome Detective dengue tool DENV-2 genotypes were also named based on geographic associations. Since dengue has spread considerably beyond the old geographic association, using only roman numerals (i.e. I, II, III, etc.) seems more appropriate. Moreover, a numeral system helps to stay in structure with new diverging lineages and genotypes. For reference, the geographic association names for DENV-2 correspond to the currently used genotype names as follows:
- Genotype I - American
- Genotype II - Cosmopolitan
- Genotype III - Southern Asian-American
- Genotype IV - Asian II
- Genotype V - Asian I
- Genotype VI - Sylvatic
Acknowledgements
Developed by: FIOCRUZ, Brazil (Vagner Fonseca, Marta Giovanetti, Maria Inês Restovic, Murilo Freire, Luiz Alcantara), KU Leuven, Belgium (Kristof Theys, Pieter Libin, Lize Cuypers, Ana Abecasis, Anne-Mieke Vandamme), University of Oxford, U.K. (Nuno Faria and Oliver Pybus), Evandro Chagas Institute, Pará, Brazil (Marcio Roberto Teixeira Nunes), CDC/OID/NCEZID (Gilberto A. Santiago), Emweb bvba, Belgium (Koen Deforche, Sara Cleemput), Krisp/UKZN, South Africa (Tulio de Oliveira), and Yale School of Public Health, USA (Verity Hill, Nathan Grubaugh).
Software: Genome Detective
Contact: Vagner Fonseca, Dr. Luiz Alcantara and/or Prof. Tulio de Oliveira